WPGP/WHISP RGL Cloning
During the past five years, progress has been made in cloning and characterizing genes from a wide range of plant taxa that confer resistance to a variety of pathogens and pests, including viruses, bacteria, fungi, and nematodes. Surprisingly, most of these genes share significant similarities in sequence homology and structural motifs. Conserved regions present in plant resistance genes allow similar sequences to be isolated from other species using homologous primers and the polymerase chain reaction (PCR) (Kanazin et al, 1996; Leister et al, 1996; Yu et al, 1996; Lagudah et al, 1996; Rivkin et al, 1996a & b). Our research employs a similar approach toward cloning the sugar pine R gene.
PCR amplification using class 1 R gene primers
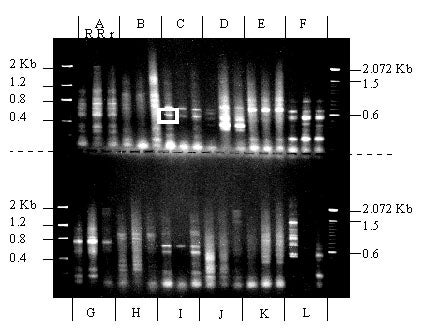
Using oligonucleotides homologous to two conserved regions (P-loop and GLPLAL) of known resistance genes and the PCR, sequences from three sugar pine megagametophytes (two R and one r) were amplified. Primer combinations A, C, D, E, F, G, and I produced clear bands of the expected size (500-600 bp) in all three samples. No differences between products from megagametophytes carrying R vs. r were detected. The band that produced clone Sp 7b3 is surrounded by a white box.
PCR amplification using class 4 R-gene primers
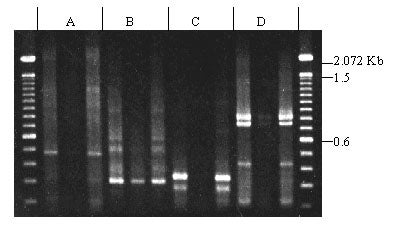
Oligonucleotides corresponding to conserved regions of the rice R gene Xa21 were used to amplify sequences from sugar pine genomic DNA. Four different kinase-domain primers were used in combination with the LRR primer, and all four primer combinations produced bands. Primer pairs A and D produced products of the expected sizes (450 bp and 800 bp respectively). No differences between megagametophytes carrying R vs. r were detected.
cloning of a RGL-fragment from sugar pine
One of the bands produced by PCR amplification using a p-loop/GLPLAL primer pair was excised and ligated into a vector, and inserts from several resulting clones were sequenced. A BLASTX sequence similarity search indicated that for one of the clones, SP 7b3, the greatest similarity was with known plant disease resistance genes. The positions of the PCR priming sites are such that the presence or absence of conserved motifs internal to the priming sites can be used for the identification of RGL sequences; these internal motifs, as well as portions of the p-loop domain internal to the primer region, are present in SP 7b3, demonstrating that a sequence related to known resistance genes is present in the sugar pine genome.
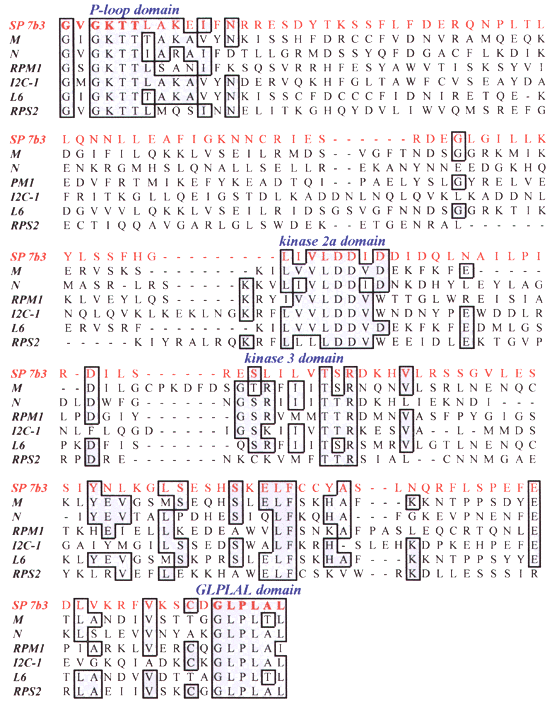
SP 7b3 is the deduced amino acid sequence of a sugar pine fragment. Other sequences are known resistance genes from various plant species. Positions at which four or more residues are identical are boxed. Conserved domains are indicated. Residues corresponding to PCR primers are in bold.
southern analysis of RGL sequences in gymnosperms
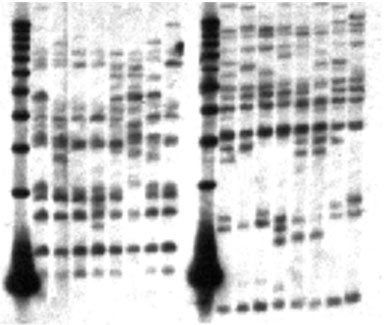
Genomic DNA from four resistant (Rr) trees and four sensitive (rr) trees was digested with restriction enzymes and probed with the cloned fragment SP 7b3. Results indicate that 10-15 copies of similar sequences are present in the sugar pine genome. Some bands are polymorphic; however, linkage with R was not apparent. Similarly, multiple fragments hybridizing to SP 7b3 were detected in the genomes of slash pine (P. elliottii), loblolly pine (P. taeda ), and Douglas fir (Psudotsuga menziesii) (not shown), indicating that RGL sequences are found throughout the gymnosperms and that RGL sequences predate the angiosperm-gymnosperm divergence. Lanes: 1, 11, 1 kb marker; 2-5, 12-15, Rr; 6-9, 16-19, rr. Lanes 2-9 were digested with EcoRI, lanes 12-19 were digested with HindIII.